
Like grep, the uniq command provides several options that modify its. print only the matching part of the line ( -o). Extract all unique geneid values found in column 9 and save to a new file called geencounts.txt. For example, the following command places the output from grep in a file called titles.The following example displays files ending in. 2) Use -f uniq option to skip the prior columns, and use the -w uniq option to limit the width to the tmpfixedwidth. To use grep as a filter, you must pipe the output of the command through grep. grep recursively ( -R) for a regular expression ( -e) 1) Temporarily make the column of interest a fixed width greater than or equal to the field's max width.Grep -R -h -o -e FormatMask=\"* * | sed 's/FormatMask="//g' | sort | uniq grep -f File1.txt base.csv >output.txt It's not clear what you hope for the inner loop to do it will just loop over a single token at a time, so it's not really a loop at all.

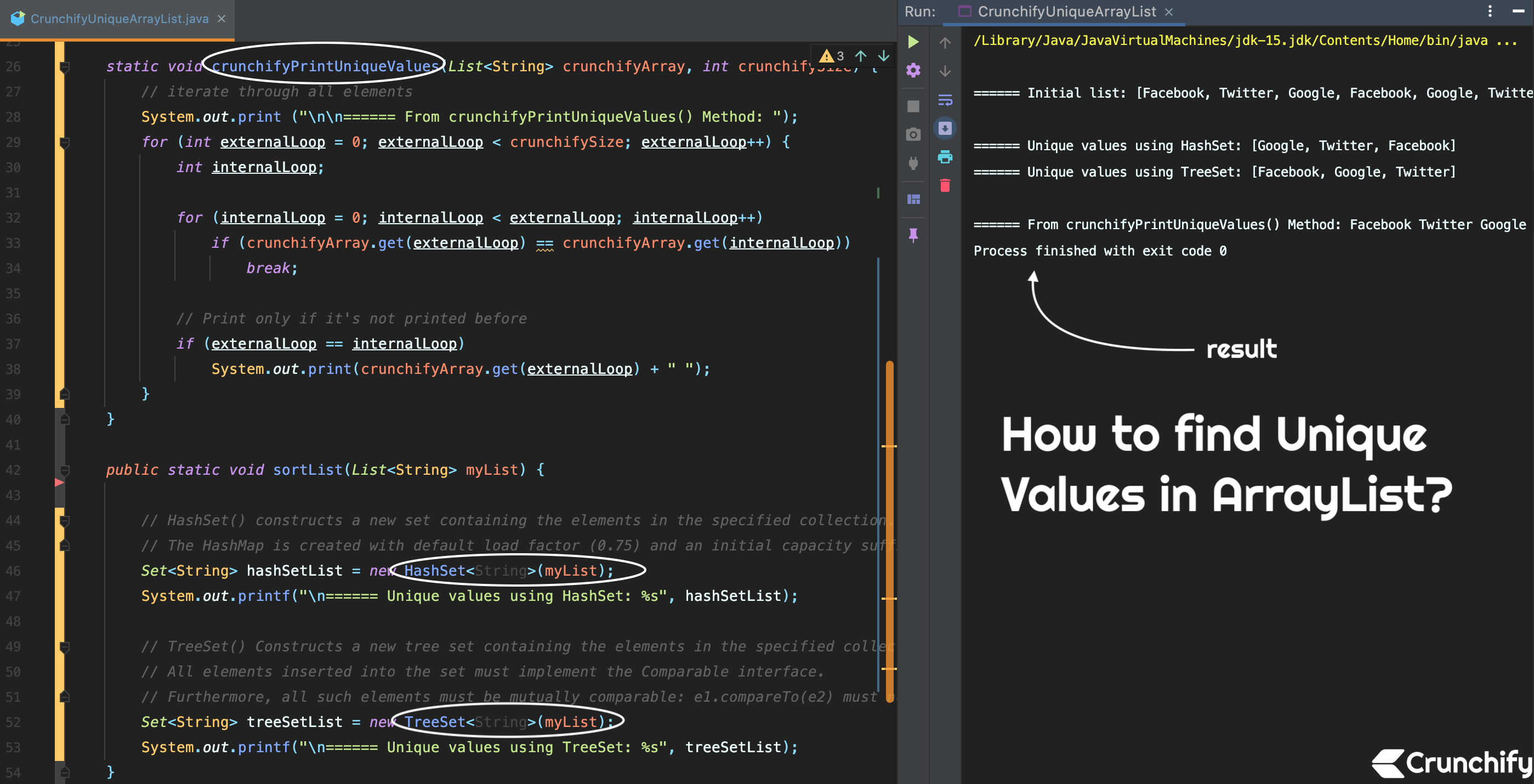
These are just 2 of the many features of the sort command. When I want to perform a recursive grep search in the current directory, I usually do: grep -ir 'string'. grep can process multiple files in one go, and then has the attractive added bonus of indicating which file it found a match in. After some iterations I came to the following command, which does what I want in a single line. Search for characters or patterns in a text file using the grep command. As you can see in the man page, option -u stands for 'unique', -t for the field delimiter, and -k allows you to select the location (key). The input looks as follows:Ī naive grep command would print out the whole line, including the file name. You can use simply the options provided by the sort command: sort -u -t, -k4,4 file.csv. END is a specially pattern that matches after all lines are processed.I have a ton of Oracle Forms XML export files and wanted to know, which different patterns occur for the value of the FormatMask XML attribute.
| awk -F, '$2 = "val1" & $3 = "val2" 'Īwk automatically trims whitespace from the values. If awk is available (it is pretty standard) you can do it like this: $ echo -e 'a,b, c,d\na,val1,val2,c' \
In case you are using git, the command git grep -h
The one marked as duplicate is different because it it is not about grep. The pipe grep "Value1" | grep "Value2" from you question does not do what you specify - it would match too much, e.g.: at 7:53 I vote for reopening this question. The first command gets the content of the File.txt file. The meta-characters ^ and $ match the beginning and the end of a line. The grep command from Linux is one of the powerful commands to find files containing some text, but when you use grep, it not only print the file name but. These commands find the number of unique words in a text file. Note that the grep pattern allows trailing whitespace after/before the values (you can remove the * sub-patterns if you don't want that). For testing purposes, the columns 2 and 3 are used instead of 14 and 15. | cut -d ',' -f2,3 | grep '^ *val1 *, *val2 *$' | wc -lĪssuming, as delimiter (and no somehow escaped, is included) in the input. If awk is not available you can do it with cut, grep and wc: $ echo -e 'a,b, c,d\na,val1 ,val2,c' \


 0 kommentar(er)
0 kommentar(er)
